Introduction: Structural variants (SVs) play a role in proliferation of plasma cells that characterizes multiple myeloma (MM). Several hallmark SVs have been identified in MM, including translocations involving the immunoglobulin heavy chain locus and several partner genes, as well as deletions and duplications affecting key genes such as TP53, RB1, and MYC. Complex SVs such as templated insertions, chromothripsis, and chromoplexy are associated with disease progression but have not yet systematically been studied.
Methods: Paired short-read (Illumina, average depth 75x), long-read HiFi (PacBio, 16x), optical genome mapping (OGM; Bionano Genomics, 341x) and Micro-C (Cantata Bio, 45x) data were sequenced from 8 relapsed MM patient-derived xenograft samples and 2 cell lines, covering t(11;14), t(4;14), and hyperdiploid subgroups. Reads were aligned to the hg38 reference genome while complex SVs (chromothripsis, chromoplexy and templated insertion) were systematically identified and accurately reconstructed by combining short-read (Manta), HiFi (pbsv), OGM, and Micro-C data (EagleC). Chromosome organization including loops, topologically associated domains (TADs) and A/B compartments were called from Micro-C data (Mustache and Juicer). Haplotype phasing was conducted using small variants detected from HiFi data. De novo assembly of the genome was conducted using HiFi and Micro-C data and compared to the reference genome. Reconstructed derivative chromosome interactions due to complex SVs were further identified by aligning Micro-C reads to the newly assembled genomes. Previously identified super-enhancers from H3K27Ac ChIP-seq from MM cell lines were mapped onto these SVs and expression of target genes assessed by RNA-seq.
Results:
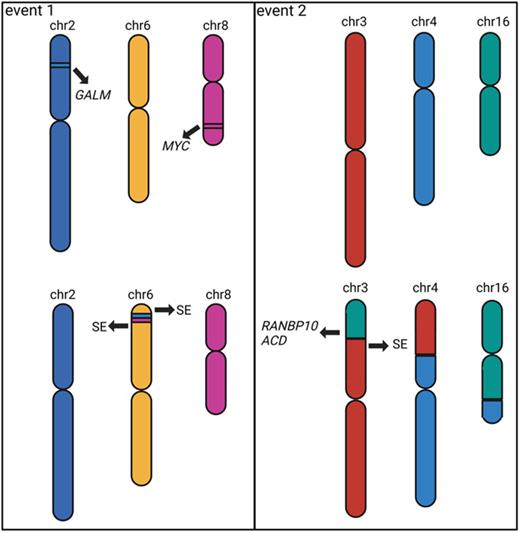
1,340 SVs were found in 10 samples using OGM of which 828 were identified in short-read WGS data. 16 complex SVs including chromothripsis, chromoplexy and templated insertions were detected in 10 samples, and we describe two (Fig.1) in detail here.
A complex templated insertion consisting of a t(2;6;8) was found in a t(11;14) PDX sample with copy number gains at breakpoints which could not be resolved from short-read data alone. OGM data revealed a single contig consisting of DNA from chromosomes 2, 6, and 8. Micro-C data revealed interactions between these chromosomes, specifically the gained regions on chromosomes 2 and 8 showed more interactions with chromosome 6 than with other chromosomes, indicating insertions into chromosome 6. As a result, both MYC at chromosome 8 and GALM at chromosome 2 were inserted into the TXNDC5 locus, containing a known MM super-enhancer, leading to the over-expression of both MYC and GALM (log 2FC=1.7 and 1.0) in this sample. A de novo genome assembly indicated neo-TAD formation encompassing the super-enhancer and both genes. In coMMpass dataset, high expression of GALM and MYC are significantly associated with inferior overall (p=0.002 and 0.05, logrank test) and progression-free survival (p=0.01 and 0.09).
Another complex templated insertion consisting of a t(3;4;16) was identified in a hyperdiploid PDX sample, again with copy number gains at breakpoints. Although the short-read data indicated an assembly similar to the previous templated insertion SV, the OGM and Micro-C data indicated a 3-way chromosome swap where the short arm of chromosome 3 was translocated to the short arm of chromosome 4, the short arm of chromosome 4 was translocated to the long arm of chromosome 16, and the long arm of chromosome 16 was translocated to the short arm of chromosome 3. Separate contigs for the t(4;16), t(3;16) and t(3;4) were confirmed by OGM data and only 2-way chromosomal interactions were seen in the Micro-C data. This complex event led to numerous oncogenes juxtaposed to super-enhancers, which caused over-expression of target genes such as ACD and RANBP10 (log 2FC=1.0 and 0.4), both of which were on chromosome 16 and are significantly associated with inferior overall (p=0.02 and p=0.004) and progression-free survival (p=0.003 and p=0.005).
Conclusion: By utilizing multiomic techniques we were able to reconstruct complex SV events, showing different mechanisms of generating templated insertion events and identifying their biological consequences. We demonstrated how these complex events could potentially facilitate tumor proliferation via engaging the super-enhancers and regulating the expression of key oncogenes.
Disclosures
Suvannasankha:Genentech: Research Funding; Bristol Meyer Squibb: Consultancy, Research Funding; Janssen: Consultancy, Research Funding; Glaxo Smith Kline: Consultancy, Research Funding; Regeneron: Research Funding. Abonour:BMS, Janssen, Takeda: Consultancy; BMS, Janssen, Pfizer: Honoraria. Walker:Bristol Myers Squibb: Research Funding; Genentech: Research Funding; Abbvie: Speakers Bureau.